Phasing
This article provides information about phasing and how it is handled on VarSome Clinical.
What is phasing?
Variant calling algorithms can identify the set of alleles present in the locus. This is what we call a genotype. Sometimes, depending on the input data and the variant caller, the genotypes can also be accompanied by the phasing information. Phasing is the process of resolving the genotypes into haplotypes, where it is determined if an allele of one genotype is on the same chromosome or different as another allele.
Phasing in VarSome Clinical
Phasing visualization is now available in VarSome Clinical.
Phasing in the variant table
- Zygosity: the zygosity icon shows if the variant is heterozygous phased or unphased.

- Phasing Group: if a variant is heterozygous phased, this column will display a number. This number can be used to retrieve all variants within the same phasing block. You can do this by clicking the
 button.
button.
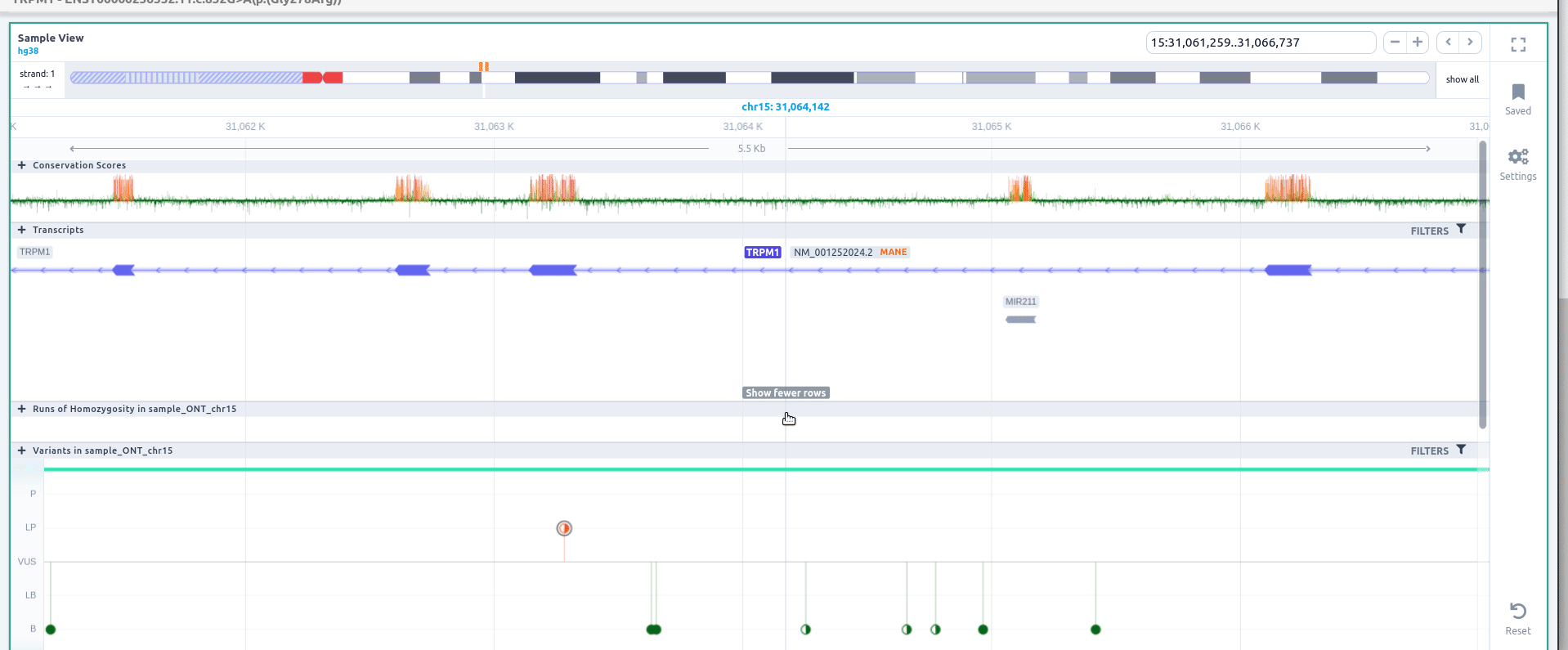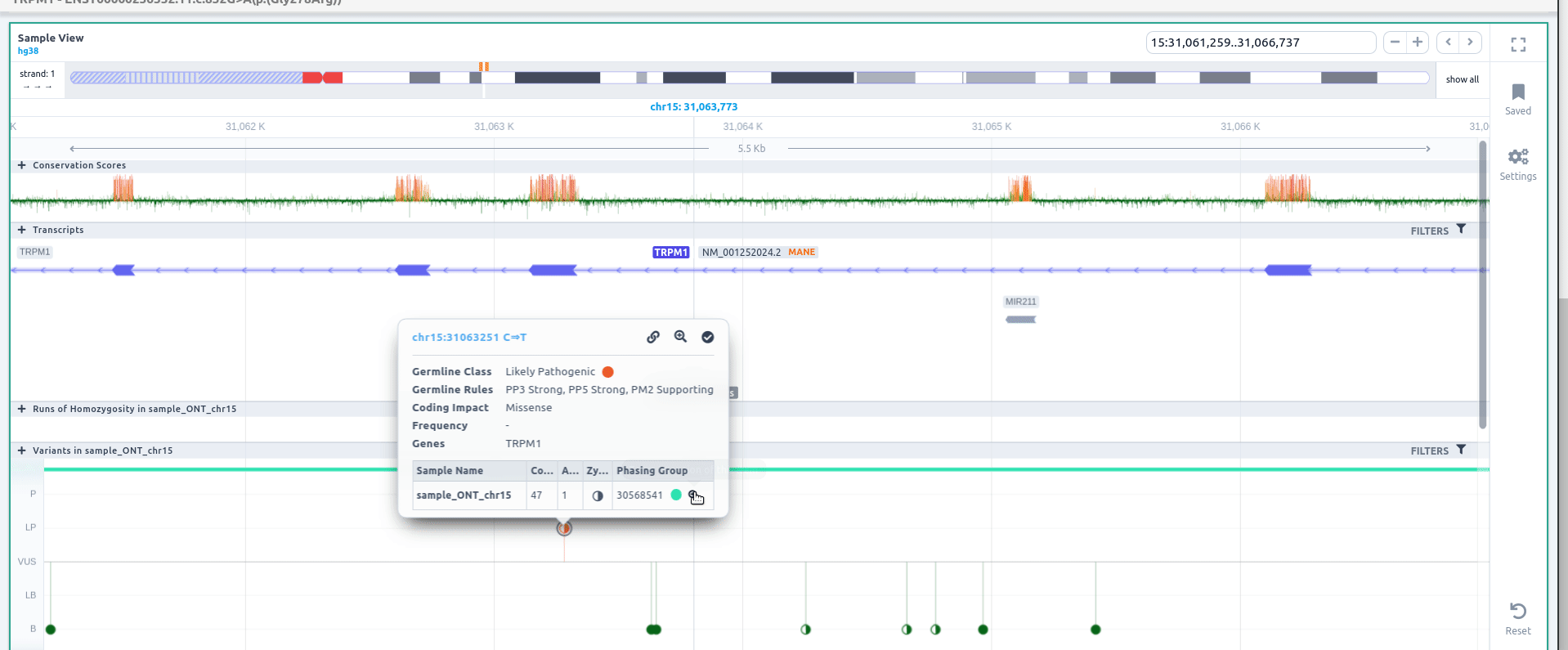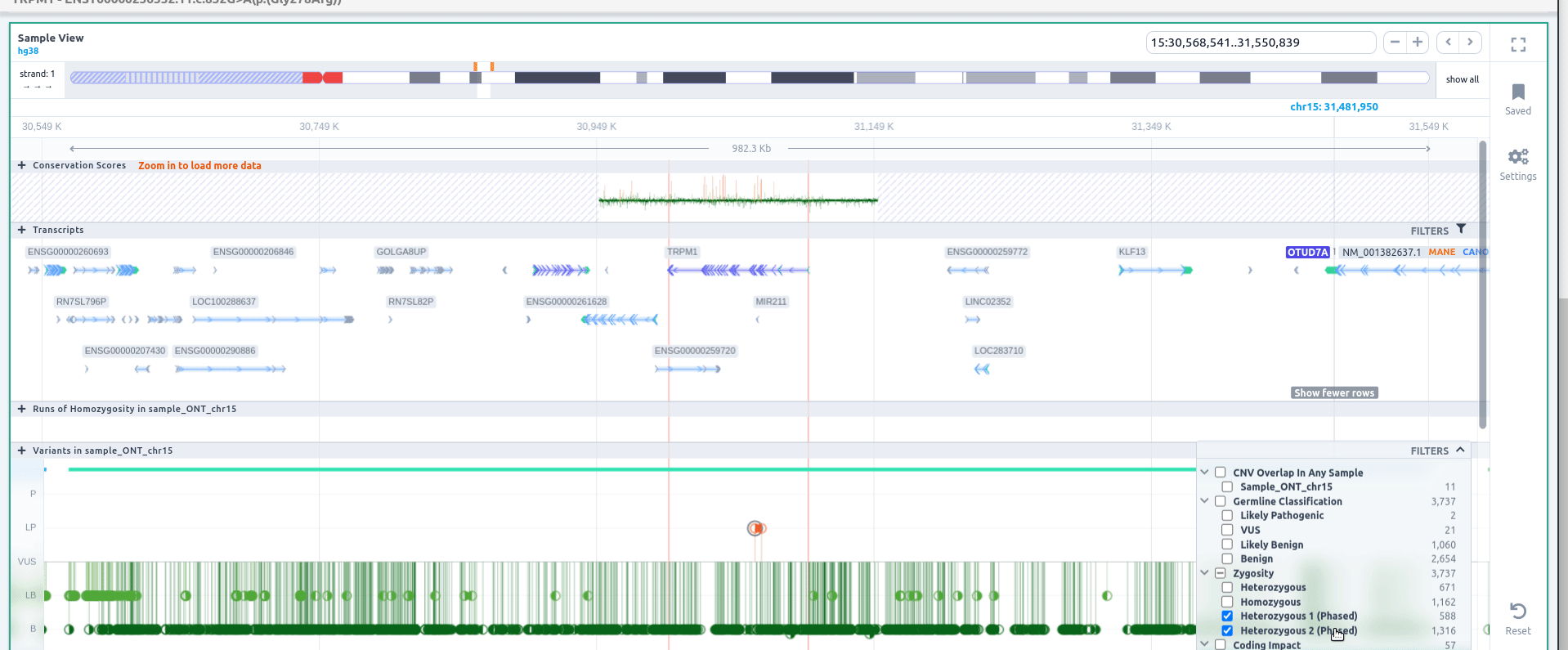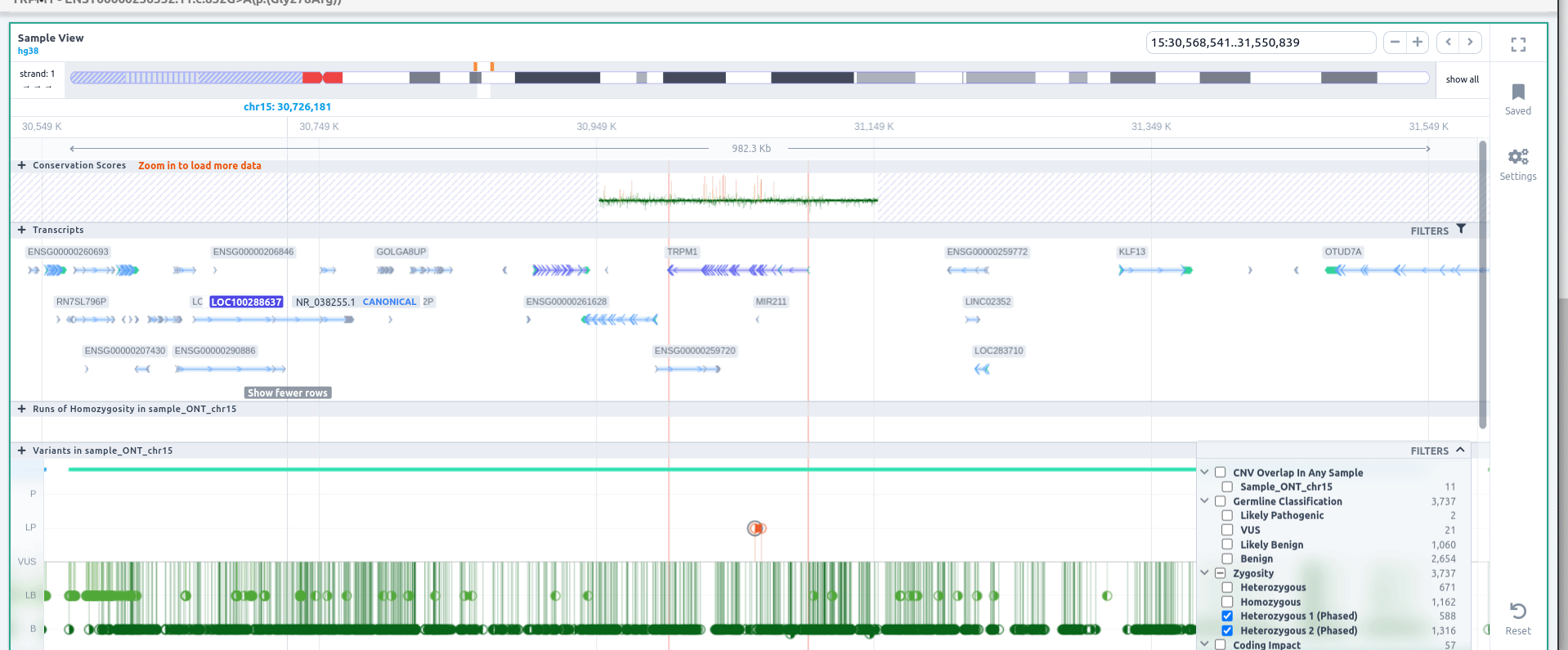
Phasing in the Sample View
The variant phasing can be also visualized in the Sample View. For more information about this feature please go here.
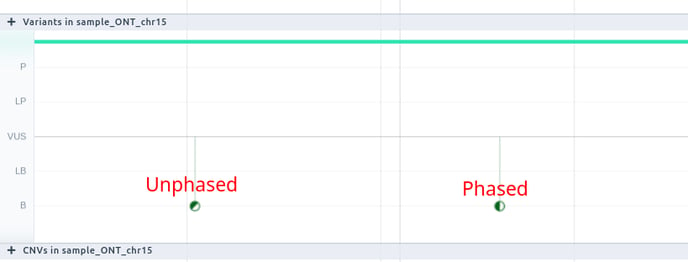
Consistently with those icons displayed in the Zygosity column of the variant table, we display whether a variant is phased or not.
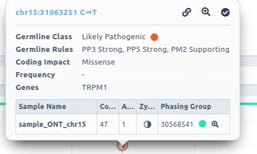
If you click on one phased variant, you can see the variant information, including the phasing group.
The phasing groups are shown above the variants
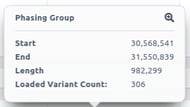
If you click on one phasing block, you can visualize the information calculated for each block: start, end, length and number of phased variants inside the phasing block.
If you click on the 🔍 to zoom when visualizing the details of a phase variant, you can zoom to the region of the block. Click on "Filters" to filter variants by zygosity, including phased or unphased variants.
How do we parse and interpret the phasing information?
The genotype field (GT) indicates if the variant contains the reference and/or the alternative allele, where 0 means the reference allele is present and 1 means the alternative allele is present. Additionally, the GT can also indicate if the alleles are phased or unphased:
- Unphased variants: if a variant is unphased (i.e. we do not know which one of the pair of chromosomes carries the variant), the genotypes are separated by a forward slash (/). E.g. ‘0/1’ stands for an unphased heterozygous variant.
- Phased variants: if a variant is phased, the genotypes are separated by a vertical slash (|). E.g. ‘0|1’ represents a phased heterozygous variant.
The genotype information of phased variants should always be accompanied by the phasing set, which is an ID of the phasing block. Otherwise, we cannot know if the caller has identified other variants phased with the variant of interest. We can only consider that two or more variants are phased if they have the same genotype AND the same phasing group/ID.
In the following paragraphs we give you some examples of how genotype and phasing information can be interpreted.
Example 1: Unphased variants
|
Position |
REF allele |
ALT allele |
Genotype (GT) |
Phasing set (PS) |
|
chr10-20,000 |
C |
A |
0/1 |
- |
|
chr10-20,050 |
T |
G |
1/0 |
- |
|
chr10-20,100 |
A |
C |
0/1 |
- |
Interpretation: the three variants reported are unphased as indicated by the forward slash symbol. This means that we do not know which allele is on which haplotype. In this case whether these variants share or not the same phasing set is irrelevant.
Example 2: Phased variants
|
Position |
REF allele |
ALT allele |
Genotype (GT) |
Phasing set (PS) |
|
chr10-20,000 |
C |
A |
0|1 |
20000 |
|
chr10-20,050 |
T |
G |
1|0 |
20000 |
|
chr10-20,100 |
A |
C |
0|1 |
20000 |
|
chr10-40,100 |
A |
G |
0|1 |
40000 |
|
chr10-40,200 |
T |
G |
1/0 |
40000 |
|
chr10-40,300 |
A |
T |
0|1 |
40000 |
Interpretation: we identify two phasing sets with IDs of 20000 and 40000:
- Phasing set 20000: there are three phased variants.
|
Position |
REF allele |
ALT allele |
Genotype (GT) |
Phasing set (PS) |
|
chr10-20,000 |
C |
A |
0|1 |
20000 |
|
chr10-20,050 |
T |
G |
1|0 |
20000 |
|
chr10-20,100 |
A |
C |
0|1 |
20000 |
The first haplotype (underlined) contains the reference alleles C and A at positions 20,000 and 20,100 respectively, and the alternative allele G at position 20,050. The other haplotype has the alternative alleles A and C at positions 20,000 and 20,100 respectively and the reference allele T at position 20,050.
2. Phasing block 2700: there are two phased variants and one unphased variant reported in this block.
|
Position |
REF allele |
ALT allele |
Genotype (GT) |
Phasing set (PS) |
|
chr10-40,100 |
A |
G |
0|1 |
40000 |
|
chr10-40,200 |
T |
G |
1/0 |
40000 |
|
chr10-40,300 |
A |
T |
0|1 |
40000 |
The first haplotype (underlined) contains the reference alleles A and A at positions 40100 and 40,300 respectively while the second haplotype contains the alternative alleles G and T at the same aforementioned positions. The variant at position 40200 is unphased so we do not know which allele is on which haplotype.