Investigating exon coverage
How to quickly check that all exons are covered?
There's no direct way of seeing gaps in an exon's coverage, but you can get partway there by using the Coding coverage report as opposed to the Region list coverage report:
Clicking on that option will bring up a popup where you can choose a gene list and then download a coverage report for the genes in that list. Please note that you will need to create the gene list before clicking on this option. To do so, click on Gene Lists in the top bar:

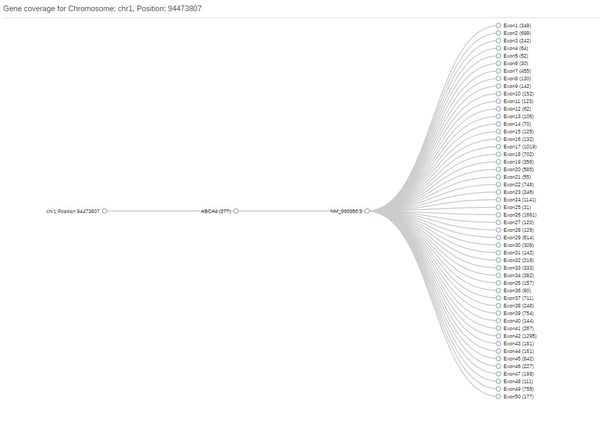
Then, click on Create new gene list and enter your genes of interest and save it with a name of your choosing. You will now be able to generate coding coverage reports using that list. This file has a breakdown of coverage by exon. Although it won't directly pinpoint gaps, it will at least show the minimum coverage per exon, so if that is 0 you know there's a gap somewhere and can then use the Gene Coverage function to locate the target exon:
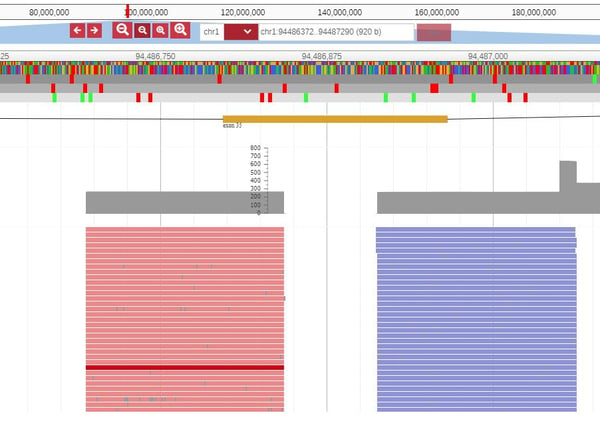
Subsequently, click on the exon and so open the JBrowse window that shows the actual reads at that position: