Microsatellite Instability (MSI) analysis on VarSome Clinical
*Research use only feature
Microsatellite instability (MSI) is a critical biomarker in cancer research, with significant implications for both therapeutic decision-making and prognosis. Microsatellites—short tandem repeats of 1 to 6 base pairs—are highly polymorphic due to their elevated mutation rates (Ellegren, 2004). These mutations primarily result from DNA replication slippage, which leads to insertions or deletions within repeat sequences. MSI is strongly associated with deficient mismatch repair (dMMR), where failures in the DNA mismatch repair system cause the accumulation of errors at microsatellite loci. Mononucleotide repeats are particularly susceptible to such errors, making MSI a powerful indicator of genomic instability and a key predictor of response to immunotherapy across various tumor types.
MSI Analysis on VarSome Clinical
VarSome Clinical assesses microsatellite instability (MSI) status using next-generation sequencing (NGS) data in somatic analyses. Unlike traditional MSI detection methods that require matched normal samples, VarSome Clinical enables accurate MSI evaluation from tumor-only sequencing data for. It supports somatic analyses starting from FASTQ with various assays, currently including:
- Whole Exome Sequencing (WES)
- Agilent SureSelect Cancer CGP Sequencing Panel
The tool outputs the MSI score (High or Not Detected) as the fraction of unstable sites (sites with evidence of indels or misalignment in microsatellite repeats) to qualifying sites (sites with adequate coverage analyzed for instability).
Where to find the MSI information in your analyzed sample
Any tumor-only somatic sample analyzed either with a WES sequencing assay (determined as assays targeting >= 31M bp that are not WGS) or an Agilent SureSelect Cancer CGP panel will get an MSI classification. In the results page of an analysis, the MSI status is visible above the Variant Table:
In addition, the details of the MSI status can be found in the Sample/Analysis Information action:
MSI Estimation Methodology
Baseline for MSI Analysis
MSI assessment is based on establishing a reference distribution of microsatellite stability using data from clinically normal samples. In VarSome Clinical, this baseline is derived from alignment files (BAM) of normal tissue from 1,532 cancer cases in The Cancer Genome Atlas (TCGA) Program, specifically from three cancer types: colorectal cancer (CRC), uterine endometrial carcinoma (UCEC), and stomach adenocarcinoma (STAD). Probability values were computed across a large number of microsatellite loci, enabling the identification of instability by detecting deviations from this reference distribution.
MSI classification
The MSI threshold serves as the cutoff distinguishing between MSI-High and MSI-Not Detected classifications, ensuring reliable and consistent MSI evaluation. Different thresholds were determined by optimizing classification performance using 80 TCGA cases with known MSI status, specific to each cancer type. For colorectal cancer (CRC), uterine corpus endometrial carcinoma (UCEC), and stomach adenocarcinoma (STAD), individual thresholds were established. For all other cancer types, a general, cancer-type-agnostic threshold was used.
- For the cancer types CRC, UCEC, and STAD, we use cancer type-specific thresholds, and the MSI status is displayed accordingly:
- MSI score >= cancer-specific threshold → MSI High
- MSI score < cancer-specific threshold → MSI Not Detected
- For any other type, a cancer-type agnostic threshold (threshold(All)) is implemented to delimit MSI high cases. An additional MSI status class is used, MSI High - equivocal for samples with MSI scores that, while lower than the threshold, are still higher than the lowest of the cancer-type specific thresholds:
- MSI score >= threshold (All) → MSI High
- Lowest threshold from validated cancer types <= MSI score < threshold (All) → MSI High - equivocal
- Equivocal is the state where the MSI score is within a range of values close to the optimal threshold, and the MSI high classification should be considered with caution
MSI score < lowest threshold from validated cancer types → MSI Not Detected
Note that thresholds are distinct for each cancer type, reference genome, and sequencing assay. The threshold of each unique combination of conditions is displayed in the MSI card and in the Sample/Analysis information MSI section.
Example cases
Example 1
- Cancer type: Colon adenocarcinoma (CAD)
- Assay: SureSelect Human All Exon V6 r2 (WES)
- MSI Score: 21.8% (3034/13946)
- MSI Status: MSI HIGH
In this case, the cancer-type agnostic threshold is used. The MSI status is marked as High, meaning the MSI score of 21.8% (3034 unstable sites out of 13946 assessed microsatellite loci) is beyond the 3.5% high confidence threshold.
Example 2
- Cancer type: Uterine endometrial carcinoma (UCEC)
- Assay: Agilent SureSelect Cancer CGP
- MSI Score: 1.2% (15/1205)
- MSI Status: MSI Not Detected
VarSome Clinical implements a UCEC-specific threshold. The MSI status of the sample is Not Detected, meaning the MSI score of 1.2% (15 unstable sites out of 1205 assessed microsatellite loci) is below the 2.4% threshold for UCEC.
Example 3
- Cancer type: Pancreatic Adenocarcinoma
- Assay: SureSelect Human All Exon V6 r2 (WES)
- MSI Score: 2.4% (224/9337)
- MSI Status: MSI High! (Equivocal)
In this case, the cancer type is not one of CRC, UCEC, or STAD, thus, the cancer-type agnostic threshold is considered for classification. Specifically, the MSI status is marked as High-equivocal, meaning that out of 9337 assessed microsatellite sites (MS), 224 sites were deemed unstable, giving an MSI score of 2.4% (224/9337). This MSI Score is within the equivocal zone (2.3-5.2%).
Conclusion
VarSome Clinical enables efficient and highly accurate detection of microsatellite instability (MSI) from tumor-only sequencing data, leveraging the MSIsensor-pro algorithm (Jia et al., 2020). This functionality allows specialists to extract critical genomic insights for cancer diagnostics and precision medicine, without the need for matched normal samples, streamlining workflows and enhancing analytical precision.
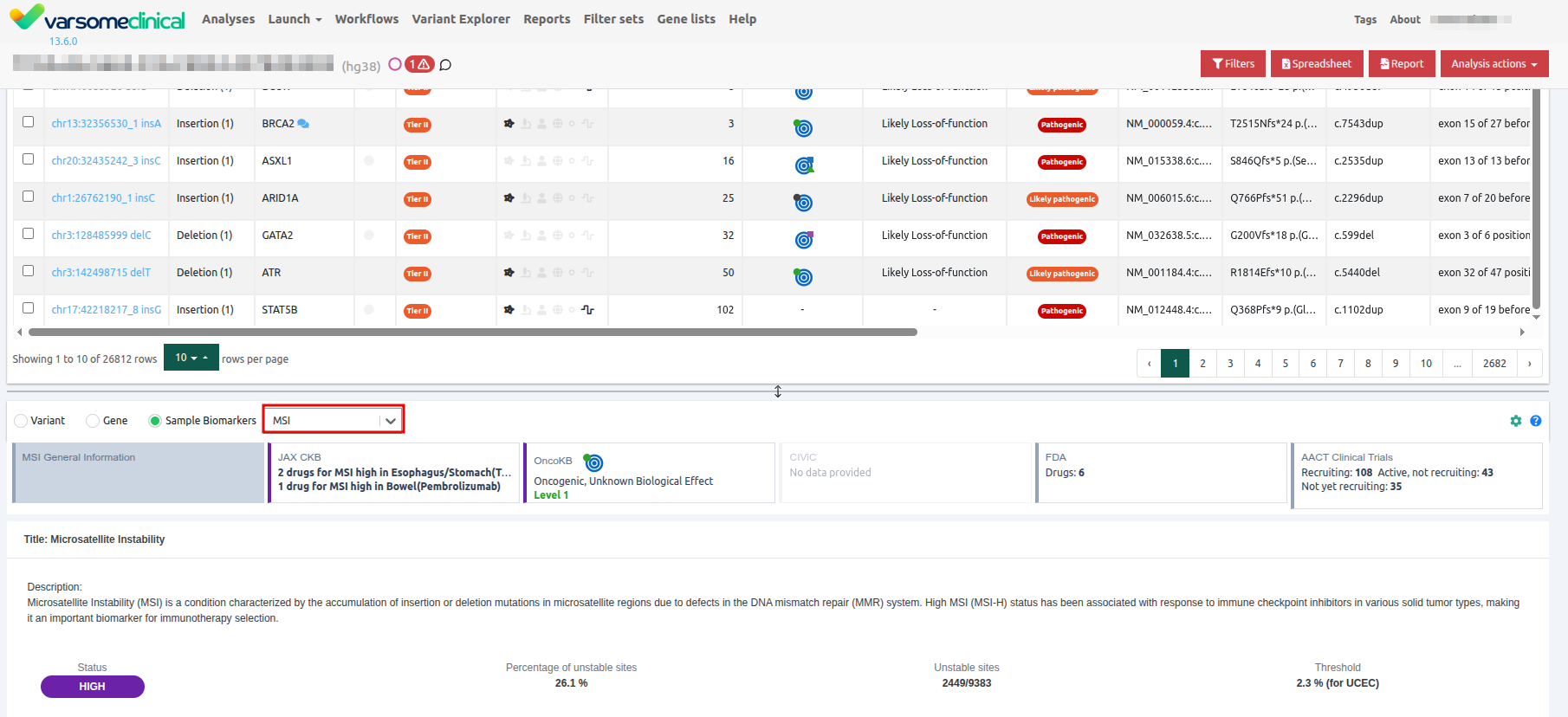
Sample Biomarker Card for MSI
You can also view the MSI car in the UI by clicking in the "Sample Biomarkers" toggle.
-png-3.png?width=688&height=330&name=image%20(3)-png-3.png)
And then selecting "MSI".
-png-1.png?width=688&height=340&name=image%20(5)-png-1.png)
You'll then be able to see the MSI General Information card.
References
Ellegren, H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5, 435–445 (2004). https://doi.org/10.1038/nrg1348
Peng J et al., (2020) MSIsensor-pro: Fast, Accurate, and Matched-normal-sample-free Detection of Microsatellite Instability, Genomics, Proteomics & Bioinformatics, Volume 18 (1): 65-71. 10.1016/j.gpb.2020.02.001


