How can I submit to ClinVar?
This article guides you through the process of submitting to ClinVar from VarSome, VarSome Premium or VarSome Clinical.
Submitting from VarSome.com
Step 1
Register on the ClinVar Submission Portal.
This requires you to provide information about the submitting organization and the people associated with that organization.
Request a service account
Submission by API requires a Service account for the submitting organization and an associated API key. Please contact clinvar@ncbi.nlm.nih.gov to request a Service account. You need to provide an email address that you want to be associated with the service account.
Both of these actions are mandatory for using this feature. Any existing VarSome user/organization must already be registered and logged in to the application in order to submit to ClinVar.
Step 2
Search for the variant of interest on VarSome starting from one of the two options (1) and Click on the ‘’Submit to ClinVar’’ button (2) available on the Variant page:
- Home page
- Variant page
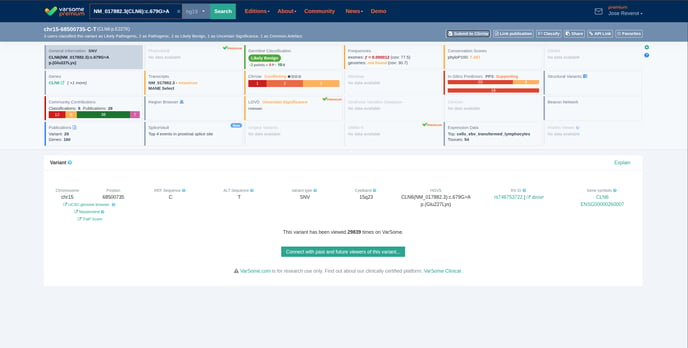
The ‘’Submit to ClinVar’’ button is available in the ClinVar card as seen on the screenshot below.
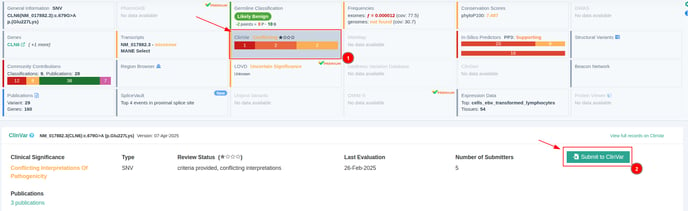
NOTE: Once you click on the submit button, if you have not previously configured the ClinVar API key, you will get a message prompting you to do so (as shown below).
Step 3
Complete the Submission form.
Assertion Criteria
In order to submit to ClinVar you will have to provide evidence describing the criteria you use to classify variants. This is called "Assertion criteria" and you can provide it through a single citation, URL or a file name.
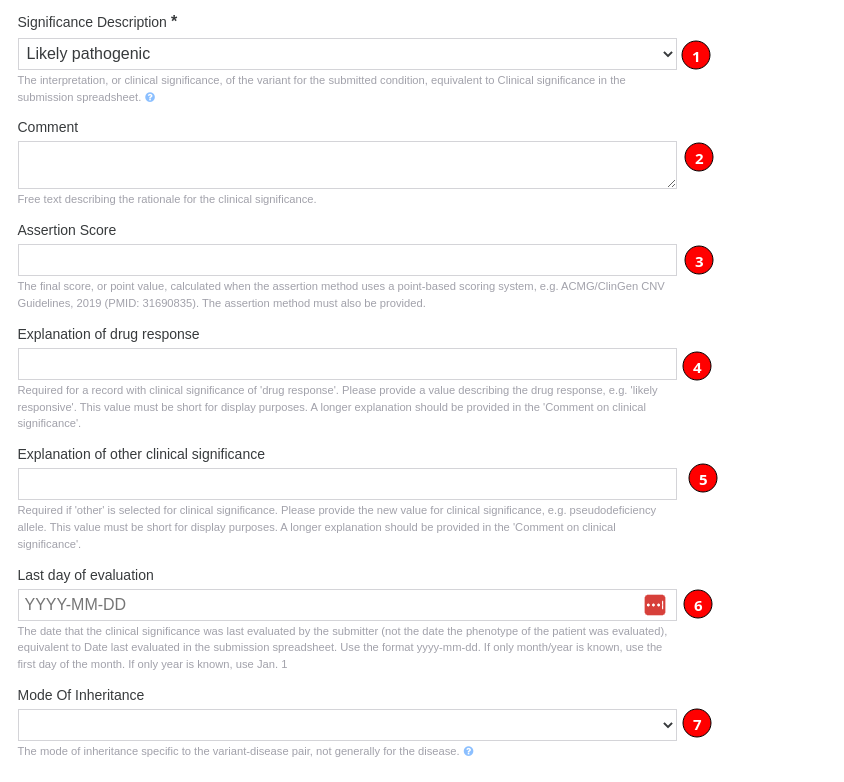
Clinical Significance
Here you reference citations that were used to evaluate the clinical significance of the variant. More than one citation may be provided (1). Each citation can be provided as a database identifier, like a PubMed ID, or a URL, but not both.
NOTE: In cases where you provide the Citation ID, and it’s origin is from PubMed, our system will validate it and display the Abstract to you.
Next, you need to select a significance description which concerns the interpretation, or clinical significance, of the variant for the submitted condition, equivalent to clinical significance in the submission spreadsheet.
You can also provide extra evidence to support the classification given with a comment (2) describing the rationale for the clinical significance (1). Additionally, you can insert an Assertion Score when the assertion method selected uses a point-based scoring system (3).
When selected "Drug response" as significance description, you can provide an explanation of drug response (4) and when selected "Other" you can use the "Explanation of other clinical significance" field (5) to provide the new value for clinical significance.
Finally, you can provide the date when the clinical significance was last evaluated by the submitter (not the date the phenotype of the patient was evaluated), equivalent to date last evaluated in the submission spreadsheet. Please use the yyyy-mm-dd format. If only month/year is known, use the first day of the month. If only year is known, use XXXX-01-01. (6). Submitter should provide a value for clinical significance in the context of a submitted mode of inheritance (7).
Drug Response
This section will be available only when "Drug Response" is selected as Significance Description in the Clinical Significance section. The condition for which the drug is used specified by database must be provided as either a database identifier (OMIM, MedGen, Orphanet, MeSH, HP, MONDO) or a name, but not both.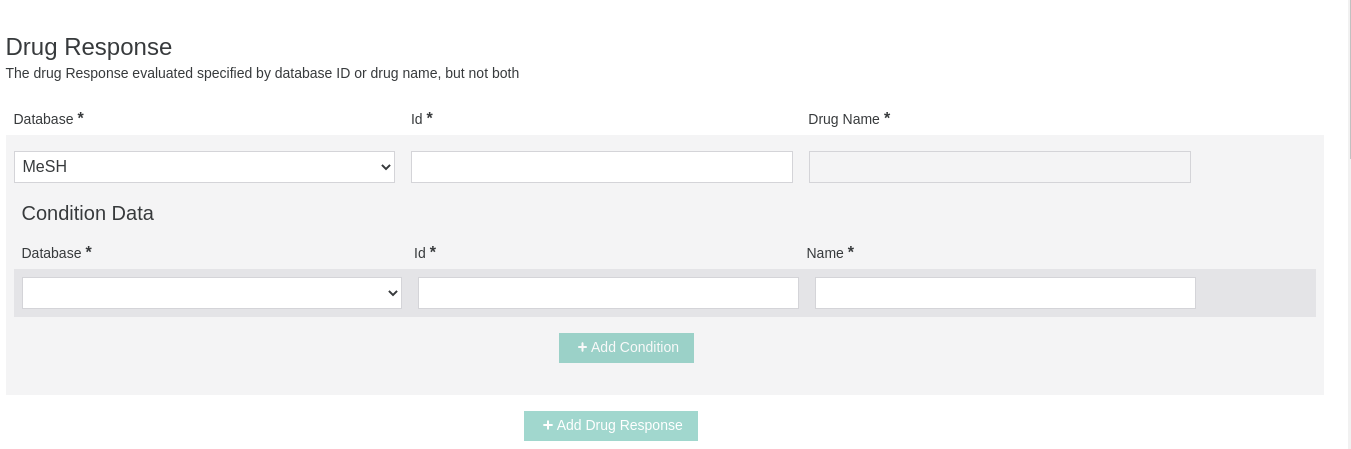
Condition(s) set
The next section concerns the condition for which the variant is interpreted. The condition must be provided as either a database identifier (OMIM, Orphanet, MedGen etc) or a name, but not both (1).
You have the option to enter the condition with the Local ID your organization uses to identify this variant or your Local Key, your unique local identifier for the variant-condition pair, equivalent to the Linking ID in the submission spreadsheet.(2)
Observed In
Additionally, it is necessary to provide supporting evidence about the affected status which Indicates whether or not the individual(s) in each observation were affected by the condition for the interpretation, as well as the allele origin(1).
If you would like to add Clinical Features please click first on ‘’Add Clinical Feature button’’.
Clinical Features that were observed by the submitter in an individual with the variant can be described in this section, optionally. More than one feature may be provided and each clinical feature may be described by a database identifier or by a name, but not both.
The collection method describes the setting in which the variant interpretation is made and it’s necessary to provide data to users of ClinVar to understand the collection method procedure (2).
You also have the option to provide a free text explanation of clinical features provided in the previous column, e.g. to describe the progression of disease or diagnosis. Please use this comment to expand on the information in 'Clinical Features'. It is suggested to mention, if possible, the total number of individuals with the variant observed by the submitter as well as the method and type of analysis used to identify a structural variant (3).
The following section is about submission name (1), record status (novel/update mandatory field)(2), and the release status (Public/hold until published)(3).
If no Submission name is provided, it will be the submission ID.
Variant Set
The interpreted variant must be described either by HGVS or by chromosome coordinates, but not both.
We currently do not support the submissions of CNVs but our intention is to release this option in the future.
Once you have provided all necessary information please click Submit. You have the option to track all your submissions in the ClinVar submissions section of your Profile.
This will display a list of all submissions made by your organization using the organization ClinVar API key. It will also allow you to track their date of submission, status or be notified of any submission errors.
There are 5 possible statuses for each submission:
- Submitted
- Processing
- Error
- Processed but not published on ClinVar
- Published on ClinVar
VarSome Clinical
Step 1
Select from the Dashboard or the Analyses page the analysis that you want.
Step 2
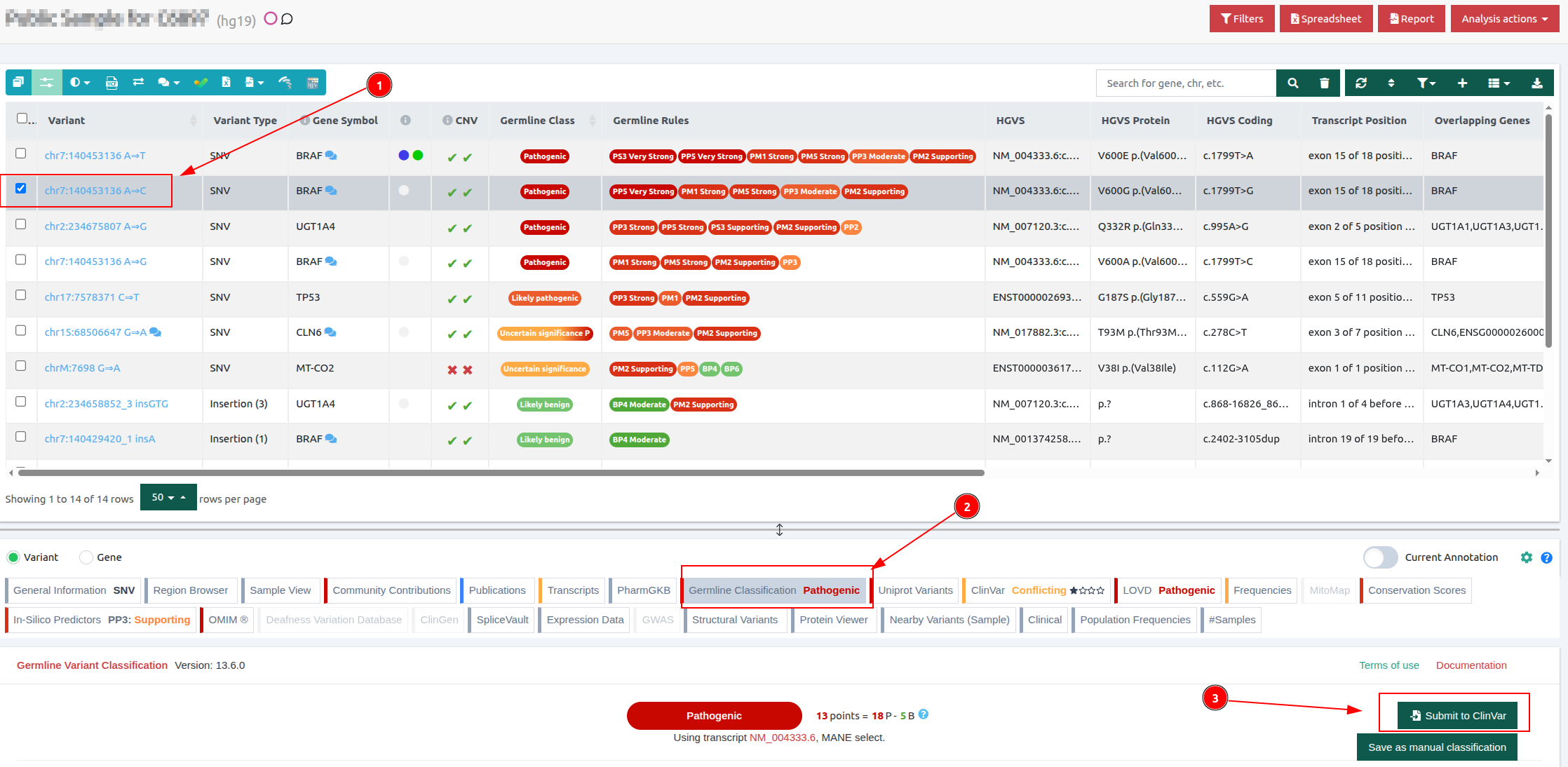
Select the variant of interest from the results table and click on the ‘’Germline Classification’’ component that is available on the VarSome Clinical results analysis page.
Then click on the ‘’Submit to ClinVar’’ button, which is available inside the Germline Classification component.
Step 3
After Submitting to ClinVar, same as in VarSome.com, you will be able to see all submissions you have made, track their status or tackle any potential submission errors.

If additional help is needed, depending on the type of the issue you might be experiencing, you can always email us at support@varsome.com or contact ClinVar directly on clinvar@ncbi.nlm.nih.gov.
-1.png?width=688&height=229&name=image%20(12)-1.png)