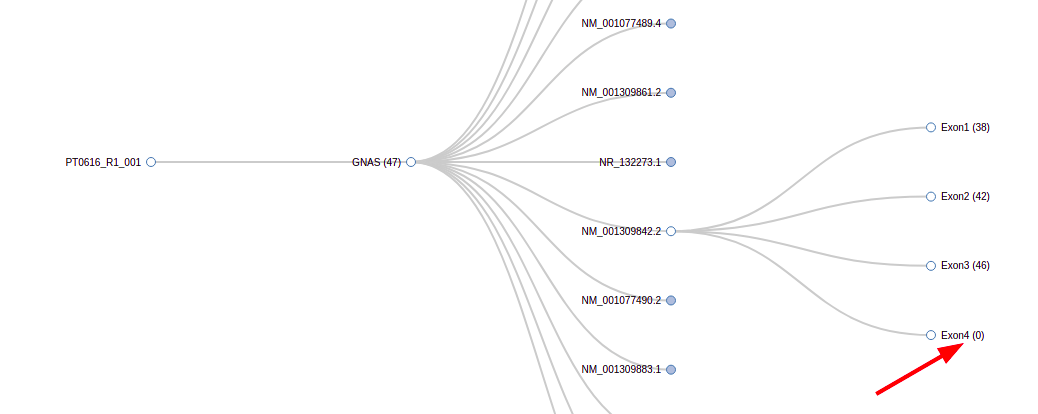
How can I quickly check the coverage for a specific exon of a given transcript or gene?
You can see exon-level coverage using our "Gene coverage" tool which you can find in the "Analysis actions" menu:

This will take you to a new page, showing a long list of genes. You can filter for your gene of interest:
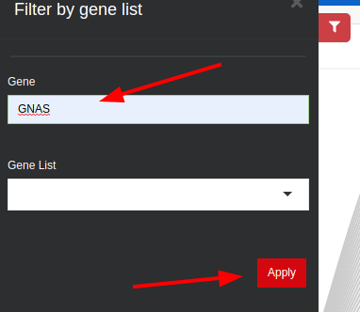
You can also access a gene's transcripts dendrogram by clicking on the "Gene Coverage" icon for a specific variant directly in the Variant Table:
When you reach the dendrogram page, you can select the transcript and exon you are interested in, and you will see the coverage for that exon in parentheses: