CNVs WGS Single-Sample analyses filtering
This article introduces a new shortcut filter designed to improve the interpretation of CNVs generated from whole-genome single-sample analyses using Delly. The filter applies specific quality and precision thresholds to exclude low-confidence and potential false-positive variants, helping users focus on high-confidence CNV calls.
When applied, the shortcut filter retains only CNVs that meet all of the following criteria:
-
Quality score (QUAL) ≥ 100, and
Sum of Split Reads (SR) and Paired-end Reads (PE) support > 6-
This criterion ensures that only variants with sufficient read support are kept.
-
Imprecise calls are typically excluded, as most lack the SR tag.
-
-
Variant length between 100 bp and 100 kb
-
Variants larger than 100 kb are often false positives and inherently pathogenic due to their size.
-
-
Precision = Precise
-
Only precise calls are retained to improve result reliability.
-
To apply the shortcut filter, simply click the filter button.
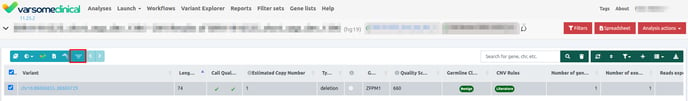
To restore the table to its original results, click the button again
In addition to the shortcut filter, you can customize results using dynamic filters for:
-
Quality
-
Variant Length
-
Precision
You can either:
-
Use the recommended thresholds listed above, or
-
Set your own parameters according to your analysis needs.
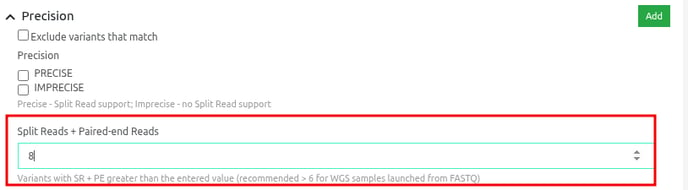
The Precision dynamic filter also incorporates the sum of Split Reads (SR) and Paired-end Reads (PE) support, allowing further customization.
The shortcut filter is only available for CNV analyses in WGS single samples launched from FASTQ.